Bacterial Communication
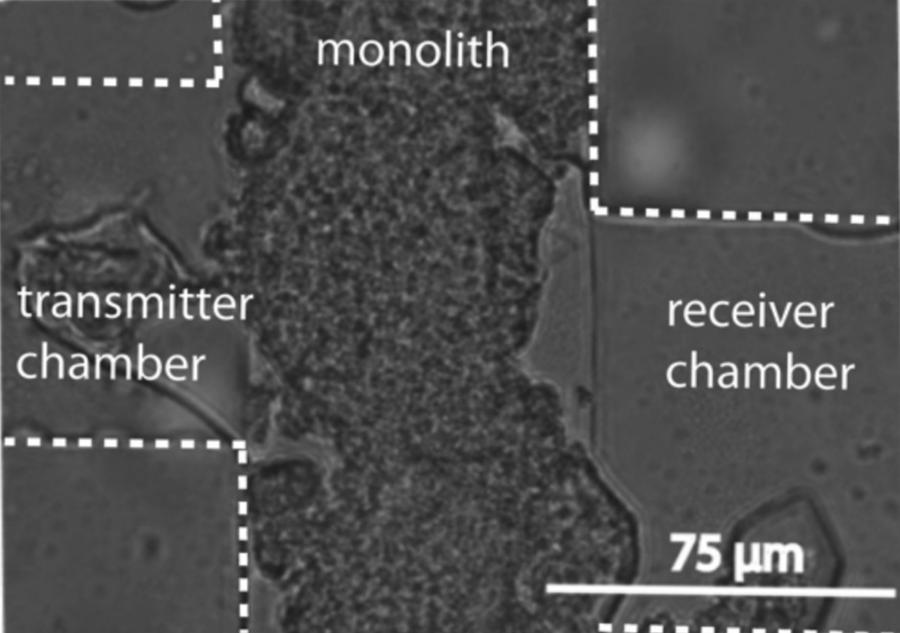
Image courtesy of Caitlin Austin
Genetically engineered bacteria can be used for a wide range of applications, from monitoring environmental toxins to studying complex communication networks in the human digestive system. Although great strides have been made in studying single strains of bacteria in well-controlled microfluidic environments, there remains a need for tools to reliably control and measure communication between multiple discrete bacterial populations. Stable long-term experiments (e.g., days) with controlled population sizes and regulated input (e.g., concentration) and output measurements can reveal fundamental limits of cell-to-cell communication. We have developed a microfluidic platform that utilizes a porous monolith to reliably and stably partition adjacent strains of bacteria while allowing molecular communication between them for several days. This porous monolith microfluidic system enables bacterial cell-to-cell communication assays with dynamic control of inputs, relatively long-term experimentation with no cross contamination, and stable bacterial population size. This system can serve as a valuable tool in understanding bacterial communication and improving biosensor design capabilities.
Related Publications
[pods name=”publication” template=”publication-listing” limit=”-1″ orderby=”date desc” where=” category.slug=’bacterial-communication’ ” ]


